Abstract
Background: Aneuploidy and large-scale Copy Number Variations (CNVs) are prominent features of cancer cells. While Fluorescence in situ hybridization (FISH) and conventional cytogenetics (CC) are the gold standard for detecting aneuploidy and CNVs, NGS-based assays are currently used for high-resolution detection of copy number alterations assessing the whole genome. However, although an increasing number of NGS-based tools have been developed for detecting aneuploidy or CNVs from whole genome or exome sequencing data, only a limited number of options are available for targeted gene panels. Despite mechanisms provided to establish normal profiles for a specific panel, the accuracy of these tools at the chromosome level suffer when only a small number of regions are targeted on each chromosome. Here we leveraged on a custom amplicon based NGS assay designed to detect somatic alterations (SNVs and indels) in 297 hematological cancer relevant genes, previously validated in our clinical laboratory. We introduce a simple approach to accurately predict chromosome-level CNVs such as monosomy and trisomy for a targeted gene panel, commonly used in a clinical setting.
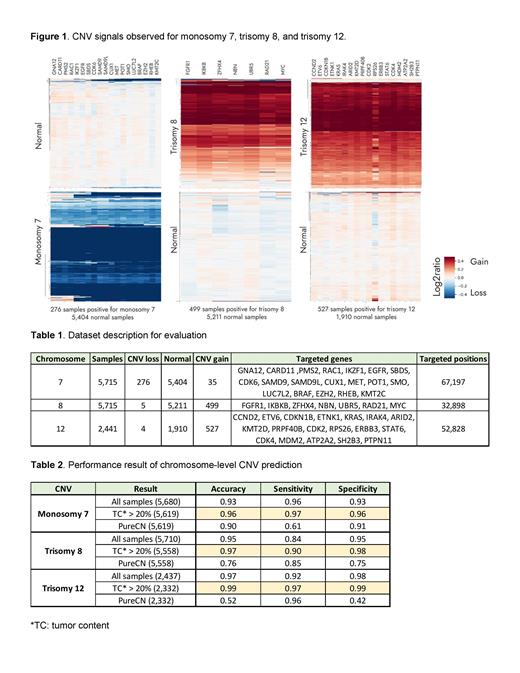
Methods: Mutation profiles, including SNVs, INDELs, and structural changes, were interrogated with an in-house bioinformatics pipeline that utilized PureCN and CNVkit algorithms to detect structural changes. The first step consists of finding optimal panel-specific decision thresholds for gains and losses at the gene level. This step was performed using an independent set of 1,314 clinical samples sequenced with the NeoType® Heme assay developed by NeoGenomics Laboratories, Inc. for which at least one FISH test was performed in addition to the sequencing. Three genes (ATM, TP53, and NF1) were used to find optimal decision thresholds based on the FISH result for these markers. These thresholds are used afterward to predict a gain or a loss for any other gene in the panel. The second step consists of predicting the chromosome-level gain or loss based on the individual predictions at the gene level by simply observing the frequency of targeted genes on the corresponding chromosome predicted as either gained or lost by the first step approach. The 19, 7, and 18 targeted genes in the NGS panel (Table 1) were respectively used to predict monosomy 7, trisomy 8, and trisomy 12 in a second set of over 7,000 clinical samples with known ploidy for chromosomes with clinically relevant ploidy abnormalities in hematological malignancies.
Results: Evaluation of the first stage gene-level CNV prediction on 1,314 clinical samples shows a concordance rate of 97.95% between NGS and FISH results on ATM, TP53, and NF1. When we evaluated the second stage chromosome-level CNV prediction in clinical samples sequenced using the same targeted panel and assessed by FISH for chromosome-level variation on chromosomes 7, 8 and 12 (Table 1), a heatmap of the predicted Log 2 ratios for each sample and targeted gene from the first step shows a clear distinctive signal between aneuploidy and diploid samples (Figure 1). At the chromosome level, the concordance rate between the final prediction and the FISH results is consistently observed above 93% (Table 2). Roughly 50% of the 12, 78, and 40 discordant calls for monosomy 7, trisomy 8, and trisomy 12, respectively captured by FISH but not by NGS can be explained by low tumor content (less than 20%) in the tested samples. The concordance rate between NGS and FISH is consistently observed above 96% when leaving these samples aside. Note that results in Table 2 are obtained using all samples to decide the optimal decision threshold for the chromosome-level prediction, but are found identical when using a leave-one-out evaluation procedure, and nearly identical when using a repeated cross-validation procedure.
Conclusion: This study demonstrates that chromosome-level CNVs can be accurately predicted in hematologic malignancies even when the number of targeted genes on a given chromosome is low. Despite the simplicity of the approach, the two stages bioinformatics pipeline based on an ensemble method allowed us to gain between 8% and 46% accuracy compared to relying only on the prediction of a single tool like PureCN. Samples with low tumor content remain, however, a difficult case to tackle with bulk NGS as it is difficult to distinguish a CNV from the natural variability of the sequencing coverage.
Nam: NeoGenomics Laboratories, Inc.: Current Employment. Magnan: NeoGenomics Laboratories, Inc.: Current Employment. Lopez-Diaz: NeoGenomics Laboratories, Inc.: Current Employment. Bender: NeoGenomics Laboratories, Inc.: Current Employment. Agersborg: NeoGenomics Laboratories, Inc.: Current Employment. Jung: NeoGenomics Laboratories, Inc.: Current Employment. Funari: NeoGenomics Laboratories, Inc.: Current Employment.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal